Background
In humans, glucocorticoids regulate a broad spectrum of physiologic functions essential for life, and they play an important role in the maintenance of basal and stress-related homeostasis. [1, 2, 3] Approximately 20% of the genes expressed in human leukocytes are regulated positively or negatively by glucocorticoids. [4] Glucocorticoids are involved in almost every cellular, molecular, and physiologic network of the organism and play a pivotal role in critical biologic processes, such as growth, reproduction, intermediary metabolism, immune and inflammatory reactions, and central nervous system and cardiovascular functions. [1, 4] Furthermore, glucocorticoids represent one of the most widely used therapeutic compounds, often used in the treatment of inflammatory, autoimmune, and lymphoproliferative disorders. [1]
Pathophysiology
Molecular mechanisms of glucocorticoid action
At the cellular level, the actions of glucocorticoids are mediated by a 94-kd protein, the glucocorticoid receptor (GR). The human (h) GR belongs to the steroid/thyroid/retinoic acid superfamily of nuclear receptors and functions as a ligand-dependent transcription factor that regulates the expression of glucocorticoid-responsive genes positively or negatively. [5, 6, 7] See panel A in the image below.
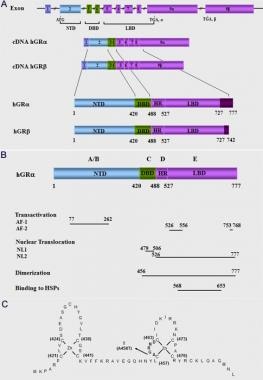 (A) Schematic representation of the structure of the human glucocorticoid receptor (hGR) gene. Alternative splicing of the primary transcript gives rise to the 2 mRNA and protein isoforms, hGR-alpha and hGR-beta. (B) Functional domains of the hGR-alpha. The functional domains and subdomains are indicated beneath the linearized protein structures. AF, activation function; DBD, DNA-binding domain; LBD, ligand-binding domain; NLS, nuclear localization signal. (C) Enlargement of part of the DBD showing the amino acid sequence (single letter codes) of the 2 zinc fingers and the dimerization loop (in bold). The A to T mutation at position 458 that could produce a dimerization defective receptor is shown.
(A) Schematic representation of the structure of the human glucocorticoid receptor (hGR) gene. Alternative splicing of the primary transcript gives rise to the 2 mRNA and protein isoforms, hGR-alpha and hGR-beta. (B) Functional domains of the hGR-alpha. The functional domains and subdomains are indicated beneath the linearized protein structures. AF, activation function; DBD, DNA-binding domain; LBD, ligand-binding domain; NLS, nuclear localization signal. (C) Enlargement of part of the DBD showing the amino acid sequence (single letter codes) of the 2 zinc fingers and the dimerization loop (in bold). The A to T mutation at position 458 that could produce a dimerization defective receptor is shown.
The hGR cDNA was isolated by expression cloning in 1985. [8] The hGR gene is one locus on the long arm of chromosome 5 (q31.3) and consists of 9 exons. Alternative splicing of the hGR gene in exon 9 generates 2 highly homologous receptor isoforms, termed α and β. These are identical through amino acid 727, but then diverge, with hGRα having an additional 50 amino acids and hGRβ having an additional, nonhomologous 15 amino acids. [3, 5, 6, 7] See the image above.
The hGRα represents the classic GR that functions as a ligand-dependent transcription factor, while the hGRβ does not bind glucocorticoid agonists; has intrinsic, hGRα-independent, gene-specific transcriptional activity; and exerts a dominant negative effect on the transcriptional activity of hGRα. [9, 10]
The human GR is a modular protein composed of distinct regions illustrated in panel B in the image below, as follows: (1) The amino-terminal A/B region, also called immunogenic or N-terminal domain (NTD) and (2) the C, D, and E regions, which correspond to the DNA-binding domain, the hinge region, and the ligand-binding domain, respectively.
 (A) Schematic representation of the structure of the human glucocorticoid receptor (hGR) gene. Alternative splicing of the primary transcript gives rise to the 2 mRNA and protein isoforms, hGR-alpha and hGR-beta. (B) Functional domains of the hGR-alpha. The functional domains and subdomains are indicated beneath the linearized protein structures. AF, activation function; DBD, DNA-binding domain; LBD, ligand-binding domain; NLS, nuclear localization signal. (C) Enlargement of part of the DBD showing the amino acid sequence (single letter codes) of the 2 zinc fingers and the dimerization loop (in bold). The A to T mutation at position 458 that could produce a dimerization defective receptor is shown.
(A) Schematic representation of the structure of the human glucocorticoid receptor (hGR) gene. Alternative splicing of the primary transcript gives rise to the 2 mRNA and protein isoforms, hGR-alpha and hGR-beta. (B) Functional domains of the hGR-alpha. The functional domains and subdomains are indicated beneath the linearized protein structures. AF, activation function; DBD, DNA-binding domain; LBD, ligand-binding domain; NLS, nuclear localization signal. (C) Enlargement of part of the DBD showing the amino acid sequence (single letter codes) of the 2 zinc fingers and the dimerization loop (in bold). The A to T mutation at position 458 that could produce a dimerization defective receptor is shown.
The NTD of the hGRα contains a major transactivation domain, termed activation function (AF)–1, which is located between amino acids 77 and 262 of the hGRα and is ligand-independent. The AF-1 plays an important role in the interaction of the receptor with molecules necessary for the initiation of transcription, such as coactivators, chromatin modulators, and basal transcription factors, including RNA polymerase II, TATA-binding protein (TBP), and a host of TBP-associated proteins (TAFIIs). [3, 5, 6]
The DNA-binding domain (DBD) of the hGRα corresponds to amino acids 420-480 and contains 2 zinc finger motifs through which the hGRα binds to specific DNA sequences, the glucocorticoid-response elements (GREs) in the promoter region(s) of target genes. [5, 6] The DBD is the most highly conserved domain throughout the steroid receptor family. The 2 zinc finger motifs are able to tetrahedrally coordinate a zinc atom and are held by 4 cysteine (Cys) residues (see the image below, panel C).
 (A) Schematic representation of the structure of the human glucocorticoid receptor (hGR) gene. Alternative splicing of the primary transcript gives rise to the 2 mRNA and protein isoforms, hGR-alpha and hGR-beta. (B) Functional domains of the hGR-alpha. The functional domains and subdomains are indicated beneath the linearized protein structures. AF, activation function; DBD, DNA-binding domain; LBD, ligand-binding domain; NLS, nuclear localization signal. (C) Enlargement of part of the DBD showing the amino acid sequence (single letter codes) of the 2 zinc fingers and the dimerization loop (in bold). The A to T mutation at position 458 that could produce a dimerization defective receptor is shown.
(A) Schematic representation of the structure of the human glucocorticoid receptor (hGR) gene. Alternative splicing of the primary transcript gives rise to the 2 mRNA and protein isoforms, hGR-alpha and hGR-beta. (B) Functional domains of the hGR-alpha. The functional domains and subdomains are indicated beneath the linearized protein structures. AF, activation function; DBD, DNA-binding domain; LBD, ligand-binding domain; NLS, nuclear localization signal. (C) Enlargement of part of the DBD showing the amino acid sequence (single letter codes) of the 2 zinc fingers and the dimerization loop (in bold). The A to T mutation at position 458 that could produce a dimerization defective receptor is shown.
Only very few amino acids, termed the proximal (P)–box, within the first zinc finger are responsible for specific recognition of the cognate GREs. Another set of amino acids, called the distal (D)–box within the second zinc finger, forms the weak dimerization interface of the DBD. The DBD of the hGRα also contains sequences important for nuclear translocation. [5, 6]
The hinge region or region D is a flexible region located between the DNA- and ligand-binding domains. Its amino terminus is an integral part of the DBD and is involved in its dimerization. The hinge region confers structural flexibility in the receptor dimmers, thereby allowing a single receptor dimmer to interact with multiple GREs. [5, 6]
The ligand-binding domain (LBD) of the hGRα corresponds to amino acids 481-777, binds to glucocorticoids, and plays a critical role in the ligand-induced activation of hGRα. The LBD also contains a second transactivation domain, termed AF-2, which is ligand-dependent, as well as sequences important for receptor dimerization, nuclear translocation, binding to the heat shock proteins, and interaction with coactivators. [5, 6]
Expressed hGRα is a panel of 8 amino terminal translational isoforms of varying lengths, each of which consists of 3 subdomains, the N-terminal (NTD), the DNA-binding (DBD), and the ligand-binding (LBD) domains. These hGRα isoforms differ at their amino-termini and may differentially transduce the glucocorticoid signal to target tissues, depending on their selective relative expression and inherent activities. It is likely that similar differential cell-specific production and functional differences might also be present between the putative hGRβ translational isoforms. [5, 6] This marked complexity in the transcription/translation of the hGR gene enables target tissues to differentially respond to circulating glucocorticoid concentrations and accounts for the highly stochastic nature of the glucocorticoid signaling pathway. [11]
In the absence of ligand, hGRα resides mostly in the cytoplasm of cells as part of a hetero-oligomeric complex, which contains chaperon heat shock proteins (HSPs) 90, 70, and FKBP51, as well as other proteins. [7, 11] Upon ligand-induced activation, the hGRα dissociates from this multiprotein complex and translocates into the nucleus, where it binds as a homodimer to GREs in the promoter regions of target genes and regulates their expression positively or negatively, depending on GRE sequence and promoter context. [7, 11] See the image below.
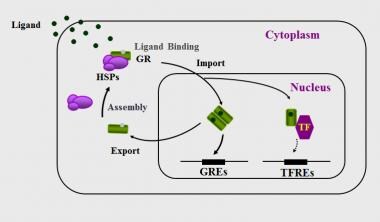 Nucleocytoplasmic shuttling of the glucocorticoid receptor. Upon binding to the ligand, the activated hGRα dissociates from heat shock proteins (HSPs) and translocates into the nucleus, where it homodimerizes and binds to glucocorticoid response elements (GREs) in the promoter region of target genes or interacts with other transcription factors (TFs), such as activator protein-1 (AP-1), nuclear factor-kappaκB (NF-kappaκB), and signal transducer and activator of transcription-5 (STAT5), ultimately modulating the transcriptional activity of, respectively, GRE- or TFRE-containing genes.
Nucleocytoplasmic shuttling of the glucocorticoid receptor. Upon binding to the ligand, the activated hGRα dissociates from heat shock proteins (HSPs) and translocates into the nucleus, where it homodimerizes and binds to glucocorticoid response elements (GREs) in the promoter region of target genes or interacts with other transcription factors (TFs), such as activator protein-1 (AP-1), nuclear factor-kappaκB (NF-kappaκB), and signal transducer and activator of transcription-5 (STAT5), ultimately modulating the transcriptional activity of, respectively, GRE- or TFRE-containing genes.
To initiate transcription, the hGRα uses its transcriptional activation domains, activation AF-1 and AF-2, located in the NTD and LBD, respectively, as surfaces to interact with nuclear receptor coactivators and chromatin-remodeling complexes. [12, 13, 14, 15] The ligand-activated hGRα can also modulate gene expression independently of DNA-binding, by interacting, possibly as a monomer, with other transcription factors, such as nuclear factor-kB, activator protein-1, p53, and signal transducers and activators of transcription. [7] Following transcriptional activation or inhibition of glucocorticoid-responsive genes, the hGRα dissociates from the ligand and has a lower affinity for binding to GREs. The unliganded hGRα remains within the nucleus for a considerable length of time and is then exported to the cytoplasm; both within the nucleus and within the cytoplasm, the hGRα may be recycled and/or degraded in the proteasome. [16] See the image above.
Alterations in the molecular mechanisms of hGRα action may lead to alterations in tissue sensitivity to glucocorticoids, which may take the form of glucocorticoid resistance or glucocorticoid hypersensitivity and may be associated with significant morbidity. [17, 18, 19] In the present review, the pathophysiology and molecular mechanisms underlying primary generalized glucocorticoid resistance, or Chrousos syndrome, are summarized.
Primary generalized glucocorticoid resistance, or Chrousos syndrome
Clinical manifestations
Primary generalized familial or sporadic glucocorticoid resistance, or Chrousos syndrome, is a rare, familial or sporadic condition, initially described and elucidated by Chrousos et al. This condition is characterized by generalized, mostly partial, target-tissue insensitivity to glucocorticoids, which leads to compensatory activation of the hypothalamic-pituitary-adrenal (HPA) axis and hypersecretion of corticotropin in the systemic circulation. [20, 21, 22] The latter results in adrenocortical hyperplasia, increased cortisol secretion as compensation for the reduced action of glucocorticoids at target tissues, and increased production of adrenal steroids with mineralocorticoid (cortisol, deoxycorticosterone [DOC], and corticosterone) and/or androgenic activity (androstenedione, dehydroepiandrosterone [DHEA], and DHEA-sulfate [DHEAS]). [20, 21, 22]
The clinical manifestations of primary generalized glucocorticoid resistance, or Chrousos syndrome, reflect the pathophysiologic alterations described above and primarily include those of mineralocorticoid and/or androgen excess. [20, 21, 22] Clinical manifestations of glucocorticoid deficiency might occur, but these are rare and have only been reported in a young child with hypoglycemic generalized tonic-clonic seizures during the course of a febrile illness [23] ; in a newborn baby with severe hypoglycemia, excessive fatigability with feeding, increased susceptibility to infections, and concurrent growth hormone deficiency [24] ; and in several adult patients with chronic fatigue. [20, 21, 22]
Clinical manifestations of mineralocorticoid excess include hypertension and hypokalemic alkalosis. Clinical manifestations of androgen excess include ambiguous genitalia in a karyotypic female at birth and gonadotropin-independent precocious puberty in children of either sex; acne, hirsutism, and hypofertility in both sexes; male-pattern hair loss, menstrual irregularities, and oligo-anovulation in females; and oligospermia in males. [20, 21, 22] The clinical spectrum of the condition is broad, ranging from most severe to mild forms, while a number of patients may be asymptomatic, displaying biochemical alterations only. [20, 21, 22]
This variable clinical phenotype is due to variations in the tissue sensitivity of the glucocorticoid, mineralocorticoid, and/or androgen receptor signaling pathways; variations in the activity of key hormone-inactivating or hormone-activating enzymes, such as the 11β-hydroxysteroid dehydrogenase [25] ; and other genetic or epigenetic factors, such as the presence of insulin resistance and visceral obesity. [21]
In recognition of Professor George P. Chrousos' extensive and ground-breaking research work in this field, it has been proposed that the term Chrousos syndrome be used in place of primary generalized familial and sporadic glucocorticoid resistance. [26, 27]
Molecular mechanisms of primary generalized glucocorticoid resistance or Chrousos syndrome
hGR mutations
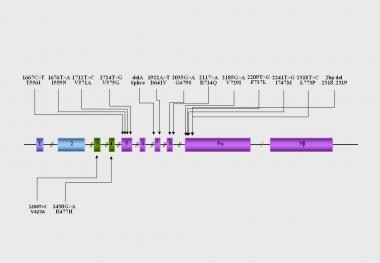
The molecular basis of Chrousos syndrome has been ascribed primarily to mutations in the hGR gene, which impair the molecular mechanisms of hGR action and decrease tissue sensitivity to glucocorticoids. [23, 24, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44] See the image below.
Most hGR mutations associated with primary generalized glucocorticoid resistance or Chrousos syndrome have been identified, and the molecular mechanisms through which these various natural hGR mutants affect glucocorticoid signal transduction have been systematically investigated in almost all reported cases of generalized glucocorticoid resistance. The following have been studied [22, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 41, 42, 43, 44] :
-
The transcriptional activity of the mutant receptors
-
The ability of the mutant receptors to exert a dominant negative effect on the wild-type receptor
-
The affinity of the mutant receptors for the ligand
-
The subcellular localization of the mutant receptors and their nuclear translocation following exposure to the ligand
-
The ability of the mutant receptors to bind to GREs
-
The interaction of the mutant receptors with the glucocorticoid receptor–interacting protein-1 (GRIP1) coactivator, which belongs to the p160 family of nuclear receptor coactivators and plays an important role in the hGRα-mediated transactivation of glucocorticoid-responsive genes
Compared with the wild-type receptor, all mutant receptors demonstrate variable reduction in their ability to transactivate glucocorticoid-responsive genes in response to dexamethasone. [28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 41, 42, 43, 44] The mutant receptors hGRaI559N, hGRaF737L, hGRaI747M, and hGRaL773P exert a dominant negative effect on the wild-type receptor, which may contribute to manifestations of the disease at the heterozygote state. [28, 32, 34, 36, 39] All mutant receptors in which the mutations are located in the LBD of the receptor show a variable reduction in their affinity for the ligand. [28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 42, 43, 44]
The only two mutant receptors that demonstrate normal affinity for the ligand are the hGRaR477H and the hGRαV423A, in which the mutations were located in the DBD. [38, 41] Most pathologic mutant receptors are observed primarily in the cytoplasm of cells in the absence of ligand, except for the hGRaV729I and hGRaF737L receptors, which are localized in both the cytoplasm and the nucleus of cells.
Exposure to dexamethasone induces a slow translocation of the mutant receptors into the nucleus, which ranges from 20-180 minutes, compared with the wild-type hGRα, which requires only 12 minutes for complete translocation. [28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 41, 42, 43, 44] All mutant receptors in which the mutations are located in the LBD preserve their ability to bind to DNA and display an abnormal interaction with the GRIP1 coactivator in vitro. [28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 42, 43, 44] The only two mutant receptors that fail to bind to DNA but display a normal interaction with the GRIP1 coactivator are the hGRaR477H and the hGRαV423A, in which the mutations are located in the DBD. [38, 41]
Epidemiology
United States statistics
Glucocorticoid resistance is rare.
International statistics
Primary generalized glucocorticoid resistance or Chrousos syndrome is rare internationally.
Sex-related demographics
Hyperandrogenism primarily occurs in children and women.
Prognosis
Morbidity/mortality
Cardiovascular morbidity and mortality is increased if primary generalized glucocorticoid resistance or Chrousos syndrome is not treated.
The syndrome should not be confused with essential hypertension.
Use of diuretics may lead to severe hypokalemia.
-
(A) Schematic representation of the structure of the human glucocorticoid receptor (hGR) gene. Alternative splicing of the primary transcript gives rise to the 2 mRNA and protein isoforms, hGR-alpha and hGR-beta. (B) Functional domains of the hGR-alpha. The functional domains and subdomains are indicated beneath the linearized protein structures. AF, activation function; DBD, DNA-binding domain; LBD, ligand-binding domain; NLS, nuclear localization signal. (C) Enlargement of part of the DBD showing the amino acid sequence (single letter codes) of the 2 zinc fingers and the dimerization loop (in bold). The A to T mutation at position 458 that could produce a dimerization defective receptor is shown.
-
Nucleocytoplasmic shuttling of the glucocorticoid receptor. Upon binding to the ligand, the activated hGRα dissociates from heat shock proteins (HSPs) and translocates into the nucleus, where it homodimerizes and binds to glucocorticoid response elements (GREs) in the promoter region of target genes or interacts with other transcription factors (TFs), such as activator protein-1 (AP-1), nuclear factor-kappaκB (NF-kappaκB), and signal transducer and activator of transcription-5 (STAT5), ultimately modulating the transcriptional activity of, respectively, GRE- or TFRE-containing genes.
-
Location of the known mutations of the hGR gene causing primary generalized glucocorticoid resistance. DBD: DNA-binding domain. GR, glucocorticoid receptor; GREs, glucocorticoid response element; HSP, heat shock protein; LBD, ligand-binding domain; NTD, amino terminal domain; TF, transcription factor; TFRE, transcription factor response element.
-
Location of the identified mutations in the hGR gene causing Chrousos syndrome.